54.1 Calibration Overview and Example Model
It is important that a model mirrors true disease progression and treatment effects as accurately as possible.
When creating the appropriate patient pathways, it can be challenging to derive appropriate Markov transition probabilities to accurately represent clinical outcomes. You may build a model based on your understanding of disease progression, but the model outputs may not match to observed data.
Calibration automatically adjusts model inputs, so that the resulting outputs match to targets. Prior to running calibration, you must do the following.
-
Setup the calibration with the appropriate optimization algorithm and tolerance levels.
-
Identify the model inputs that can be adjusted.
-
Identify model outputs to match to target values.
The calibration process then adjusts the model input values iteratively to match outputs against your targets.
Calibration Example Model
Consider the Healthcare Example Model, Calibration-Markov.trex. This Markov model is set up to run Calibration, but this chapter will take you through the steps to set up any model type for calibration.
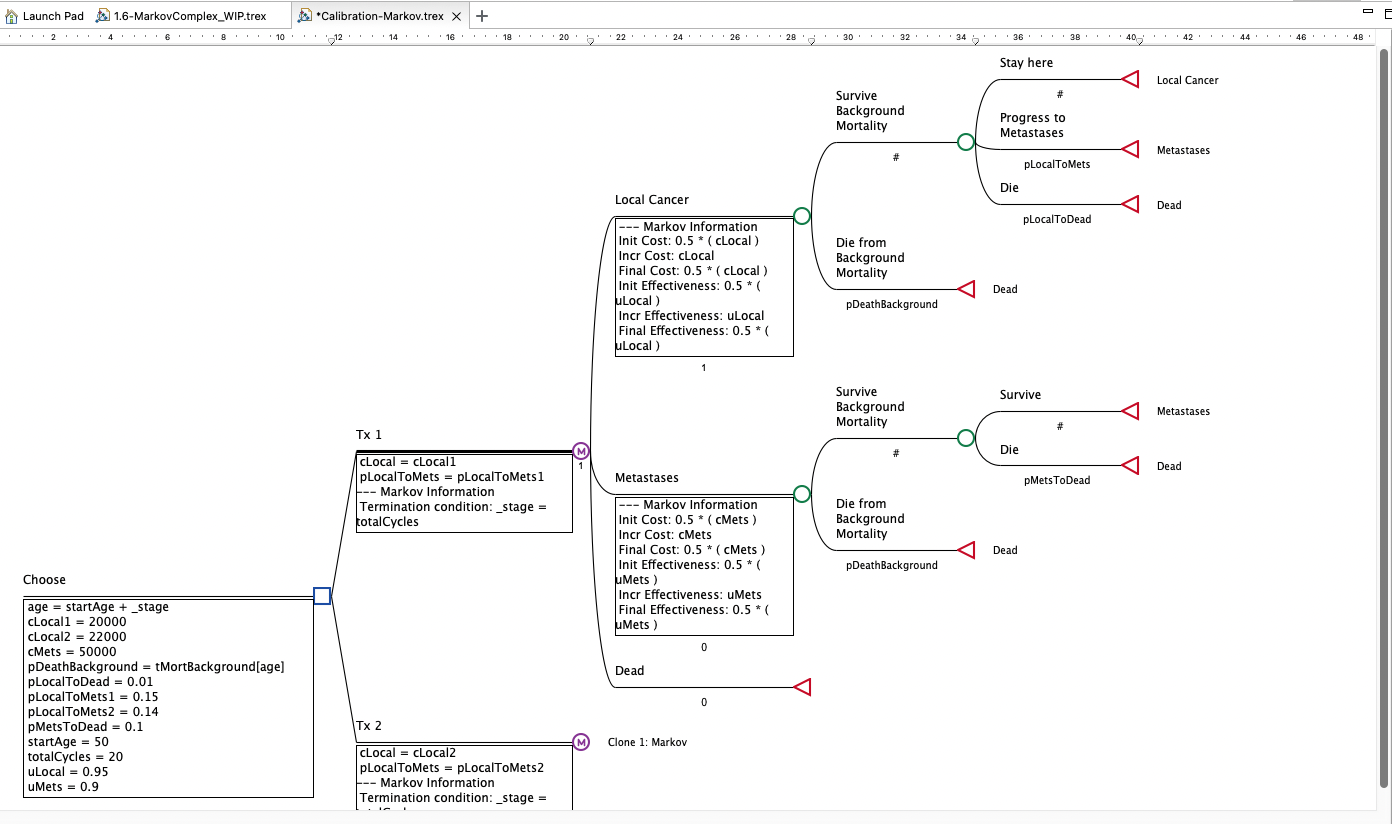
This model compares two treatment strategies - Tx1 and Tx2 - with each represented by a Markov model. The Markov model structure is the same for both treatments, with Tx1 being a clone master and Tx2 a clone copy.
We have estimated the following transition probabilities for movement between the health states.
-
Local Cancer to Metastases (pLocalToMets1 and pLocalToMets2)
-
Local Cancer to Dead (pLocaltoDead)
-
Metastases to Dead (pMetsToDead)
We also have observed 5-year progression-free and overall survival data for both strategies. When we examined the 5-year survival data generated from the model via the Markov Cohort reports, we found that it was too high.
Now let's look at how calibration helps us bring model results in line with observed data.
