39.2 Introducing Heterogeneity via Patient Characteristics
In a Microsimulation model, each patient can have his/her own set of characteristics. You would only add these characteristics into the model if they will have an impact on one or more values within the model.
There are two methods to introduce heterogeneity into the model:
-
Assigning patient characteristics by creating distributions
-
Assigning patient characteristics from real world data via Bootstrapping
39.2.1 Assigning Patient Characteristics via Distributions
Characteristics of the heterogeneous population can be provided by distributions that are sampled for each patient as he/she enters the model. TreeAge Pro supports many distribution types. Please refer to the section Creating and referencing distributions for more details.
Open the Healthcare tutorial example, MicrosimulationCancerModel - Distributions.trex, and examine the distribution: distStartAge.
The distribution, distStartAge, provides the characteristic of start age for each individual when he/she enters the model. Each patient’s start age is sampled from a Uniform distribution as a value between 30 and 50 inclusive. Due to the characteristics of the Uniform distribution, we would expect approximately the same number of people at each starting age within the range (30 - 50).
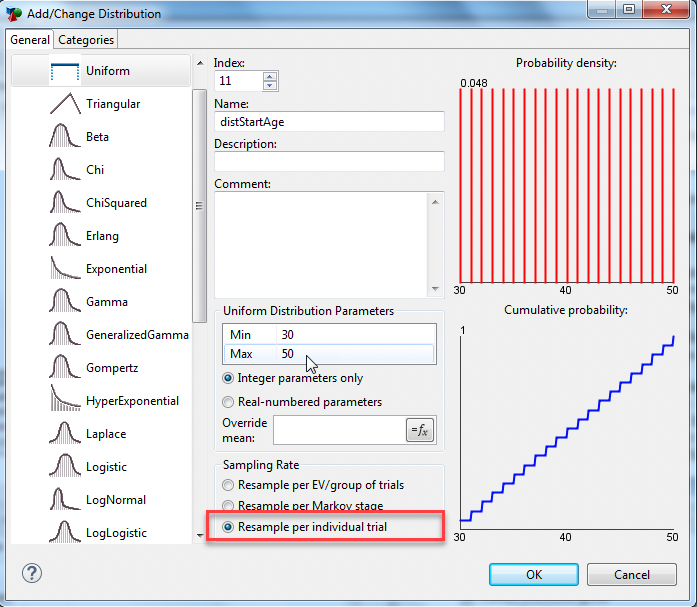
The sampling rate is set to “Resample per individual trial”, which tells TreeAge Pro to generate a new sampled start age for every person entering the model.
It is likely you will want to create distributions to match the characteristics of the cohort that experiences the disease being modeled. For example, if you have a population that affects men more frequently than women, you will want to assign patient gender based on the distribution of gender in the disease cohort.
Within the model, you can refer to the distribution directly by name. When a patient reaches a node where the start age is needed, the sampled starting age for that patient will be returned.
Refer to the section Distributions: creating, editing and using for information on creating and managing distributions.
39.2.2 Assigning Patient Characteristics via Bootstrapping
Characteristics of the heterogeneous population can be populated using real-world patient data via bootstrapping. This has an advantage over distributions in that the full set of patient characteristics is tied to a real person, so the appropriate data correlations are maintained.
Bootstrapping is implemented in TreeAge Pro with the following steps:
-
Load the real-world data into a table with each row representing a real patient and each column representing a different patient characteristic. The index column of the table should be numbered in ascending order (1, 2, 3… N, where N is the last row in the table).
-
Create a uniform distribution that returns only integers between 1 and N.
-
Create variable definitions (or tracker modifications) which assign each characteristic by pulling data from the table.
-
The table row would come from the distribution defined above (an integer between 1 and N).
-
The table column is a fixed integer for each patient characteristic.
-
The same technique defined above setting patient characteristics equal to variables can also be implemented with Trackers. The next section introduces Trackers and this current example is repeated using trackers instead of variables.
The differences between using variables and trackers:
-
If you use variables, you can define each variable at the root node and reference it anywhere in the model.
-
If you use tracker modifications, you cannot place them at the root node, so you will need to place them at each branch of the root node. Trackers should be used if you want the given characteristic values included in the Microsimulation results (i.e. the output). Doing this would allow you to filter the results by subgroup based on those characteristic values.
Open the Healthcare tutorial example, MicrosimulationCancerModel - Bootstrap With Variables.trex, and examine the distribution distProfile. The distribution generates a new profile number from a Uniform Distribution.
In the table, tProfileData[], each column is assigned a different patient characteristic, in this case tumor type (Col 1) and start age (Col 2). The profile number is used to reference a row in the table tProfileData.
The model uses the entries of the table to populate variables (or trackers) in the model for a given trial. Consider the variables defined at the root node:
-
v_StartAge = tProfileData[distProfile;2]
-
v_TumorType = tProfileData[distProfile;1]
For more details on sampling from tables refer to the section Distributions: creating, editing and using.
